How is Everyone’s Euglena Analyzed? – Part 3: Reading the DNA Sequence
How are the euglena samples, collected and sent by people from various places, analyzed in the laboratory of the Microalgal Production Control Technology Research Team?
The procedure is divided into four main steps:
- Cultivating the samples sent to us,
- Isolating them using a cell sorter,
- Reading the DNA sequence of each cultivated euglena candidate, and
- Comparing the read DNA sequences. In this article, we will introduce the reading of the DNA sequence of each euglena candidate.
How Do We Determine Something is "Euglena"?
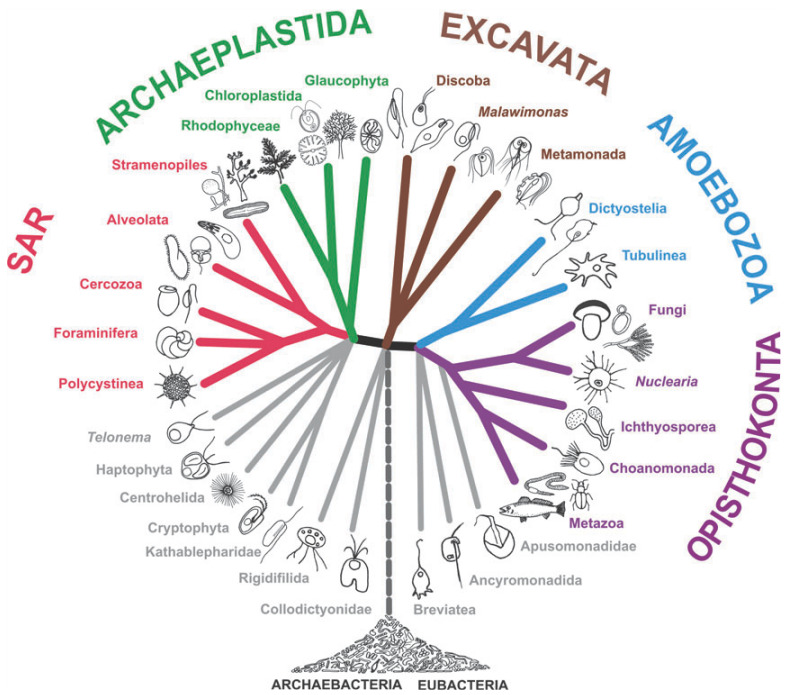
We humans classify various organisms by naming them. Euglena, especially Euglena gracilis, is classified under EXCAVATA (Reference 1). With so many organisms, how do we determine which ones belong to the same group? A traditional method is morphological observation, determining based on appearance. The samples you've sent have been observed multiple times throughout the cultivation and isolation processes to confirm the proliferation of cells that seem to be euglena.
Phylogenetic Tree

Figure 1: Phylogeny of Eukaryotes (Cited from Reference 1)
The E.gracilis species we are targeting belongs to the Euglena genus, which is believed to comprise over 200 species. Figure 2 displays photos of organisms not only from the Euglena genus but also from other genera within the Euglenales order (Reference 2). It's quite challenging to immediately determine which one belongs to the Euglena genus. Moreover, it's known that E.gracilis can change its morphology (appearance) based on cultivation conditions (Reference 3). Properly identifying E.gracilis based on appearance might require an expert eye.
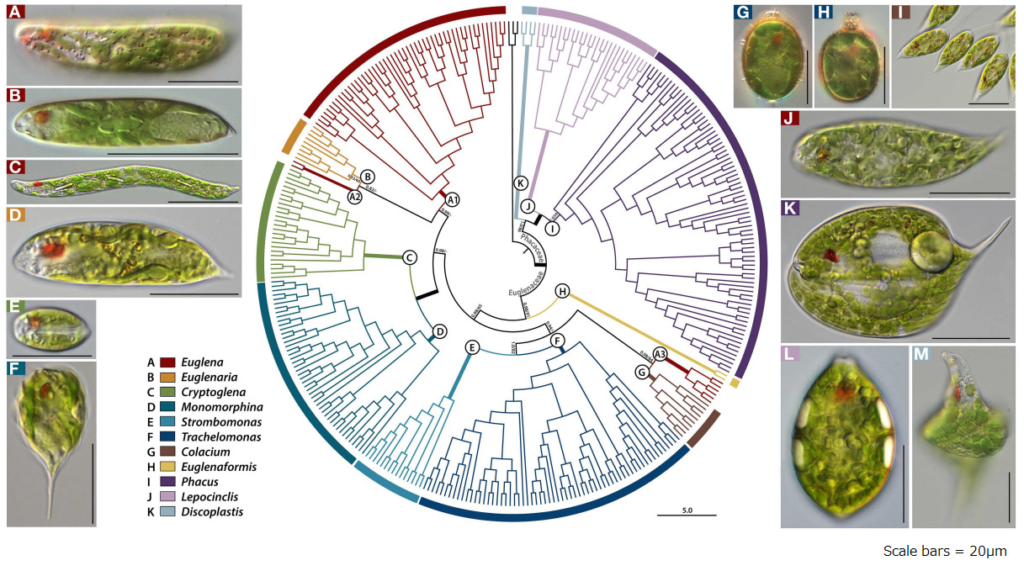
Figure 2: Members of the Euglenales order (Modified from Reference 2)
Determining Not Just by Appearance but Also by "DNA"
Of course, the Microalgal Production Control Technology Research Team has members with the expertise to identify E.gracilis. However, what we want to find from the water samples you've sent are new strains of E.gracilis, which might not look exactly like the E.gracilis we know. This is where "DNA sequencing" comes in. By reading well-preserved DNA sequences, it's possible to classify organisms as the same or different species. With the advancement of molecular biology, many organisms traditionally classified by appearance are now classified by their DNA sequences (Reference 4).
For instance, ribosomal RNA (rRNA) constitutes ribosomes that play a crucial role in protein synthesis (translation). Because of its importance, the DNA (rDNA) coding for rRNA and its surrounding DNA sequences are well-preserved across species. Comparing such DNA sequences provides another criterion to determine whether the obtained euglena candidates are the target E.gracilis.
In this article, we introduced why we read the DNA sequence of each euglena candidate as analysis step 3. In the next step (4), we will compare the read DNA sequences.
Note: This article was originally posted in June 2020 and was revised on March 1, 2022.
References
- Adl, S. M. et al. The revised classification of eukaryotes. J. Eukaryot. Microbiol. 59, 429–514 (2012).
- Kim, J. I. et al. Taxon-rich multigene phylogeny of the photosynthetic euglenoids (Euglenophyceae). Front. Ecol. Evol. 3, (2015).
- Ogbonna, J. C. et al. Heterotrophic cultivation of Euglena gracilis Z for efficient production of-tocopherol. Journal of Applied Phycology vol. 10 (1998).
- Kim, J. I. et al. Multigene analyses of photosynthetic euglenoids and new family, phacaceae (euglenales). J. Phycol. 46, 1278–1287 (2010).
