How is Everyone’s Euglena Analyzed? – Part 2: Isolation
How are the euglena samples, collected and sent by people from various places, analyzed in the laboratory of the Microalgal Production Control Technology Research Team?
The procedure is divided into four main steps:
- Cultivating the samples sent to us,
- Isolating them using a cell sorter,
- Reading the DNA sequence of each isolated strain, and
- Comparing the read DNA sequences. In this article, we will introduce the isolation using a cell sorter.
Euglena Cultivation and Isolation
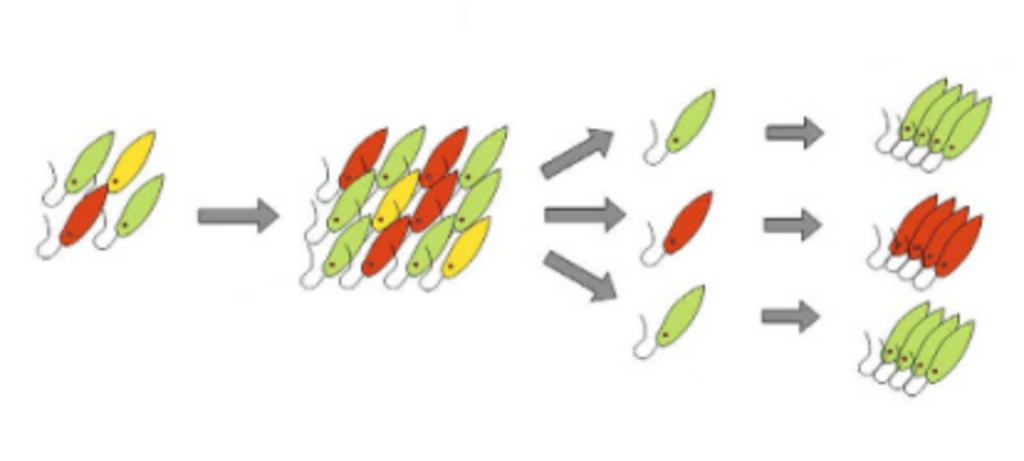
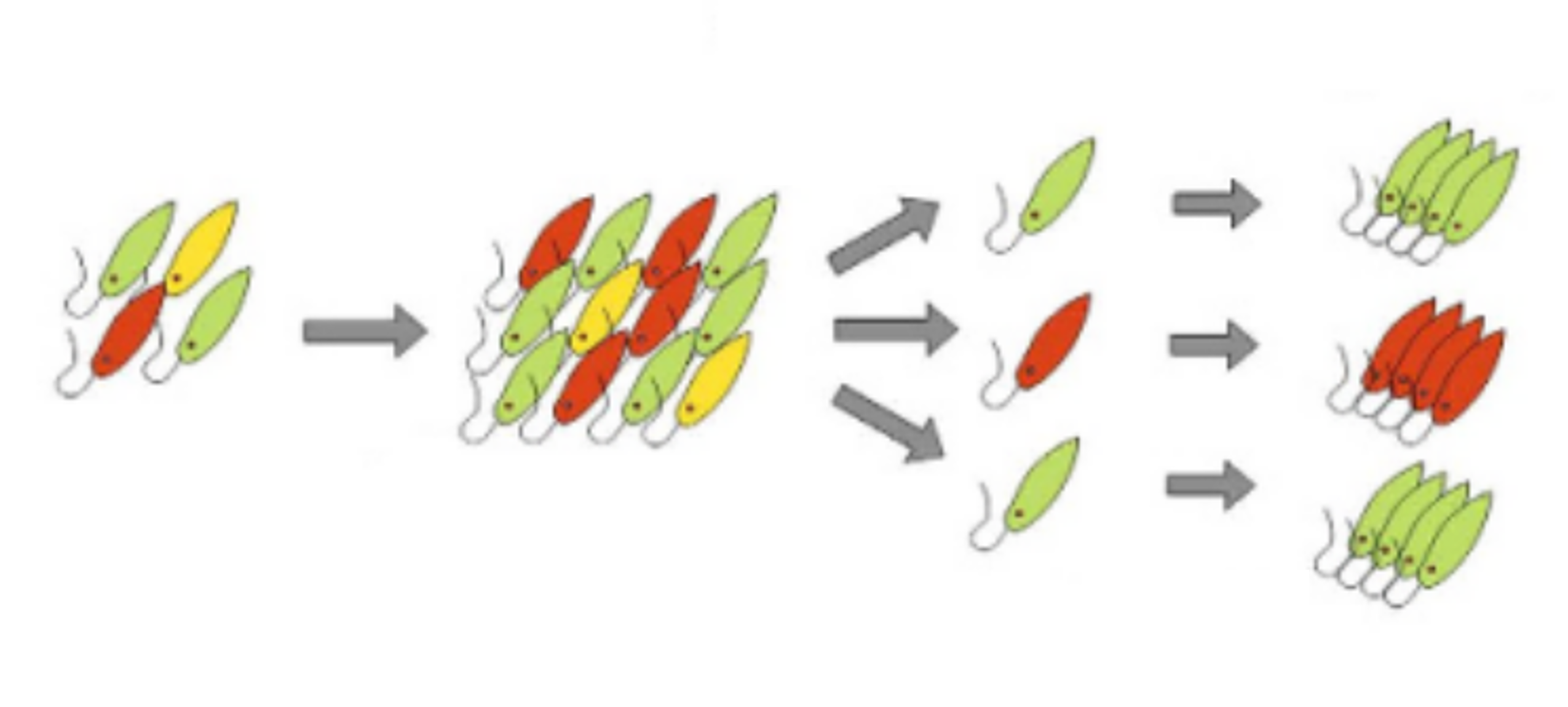
Figure 1: Cultivation and Isolation (Modified from Reference 1)
While the "euglena candidates" that survived under stringent cultivation conditions are promising, the cells proliferating in the medium might not be of a single type. Therefore, the growing cell population is processed through a device called a cell sorter, which isolates cells based on their characteristics (Figure 1). The isolated cells are then re-cultivated, and the resulting populations are referred to as "strains". Each strain undergoes morphological observation and the processes from step 3 onwards.
How Do We Distinguish Euglena?
If the goal is merely to separate a homogenous cell population, it's possible to dilute the cells so that each container receives one cell. However, the samples from the "MinMid PJ" project, even though selected in acidic media, are mixed "euglena candidates" containing various cells and bacteria. What we desire are euglena. So, how do we "distinguish" and isolate euglena from this mix?
One method is to differentiate based on morphological features like cell shape, color, and movement. Under a microscope, cells with the desired characteristics are identified and separated using a thin tube called a capillary. However, it's challenging to isolate many cells at once using this method.
FACS
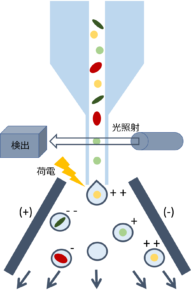
Figure 2: Cell Sorting
The cell sorter used for isolation can separate up to 70,000 droplets (liquid particles containing cells) per second at its fastest. How does it distinguish and isolate cells so quickly? The answer lies in "light" and "charge". Inside the device, each cell is streamed in a specific direction and illuminated. The light reflected off the cells is detected and analyzed to determine the cell's characteristics (flow cytometry). Based on the results, desired cells are separated in real-time according to their charge (cell sorting) (Figure 2).
In flow cytometry, the light passing through the cells reveals their size and shape, while the reflected light provides insights into the cell's internal complexity (e.g., granularity). Additionally, target proteins or lipids inside the cell can be fluorescently labeled, allowing for the determination of the presence of specific substances. The euglena E. gracilis that we target has a cell size of about 50 μm, is elongated, contains paramylon granules and chloroplasts (which emit autofluorescence, Reference 1), and produces labelable lipids under oil-producing conditions. These parameters enable us to distinguish such euglena cells.
Cells passing through the cell sorter eventually end up in individual droplets. At this point, droplets containing cells identified as "euglena" are charged. For instance, droplets charged with "+" are attracted to "-" plates, separating them from other droplets for collection.
Techniques for efficiently separating target cells are not limited to euglena and are being developed as essential tools in medical and biological fields. Besides the methods mentioned above, there are techniques like flowing cells through microfluidic devices created on substrates based on cell shape (Reference 2). There are also technologies to distinguish substances in cells without fluorescence labeling (Reference 3) and techniques using deep learning to instantly judge and separate cells based on images (Reference 4).
After isolation and re-cultivation, each strain undergoes morphological observation under a microscope (for the morphology of established strains, refer to the MinMid PJ results report). In the next step (3), the DNA sequence of each strain will be read.
References
- Yamada, K. et al. Efficient selective breeding of live oil-rich Euglena gracilis with fluorescence-activated cell sorting. Sci. Rep. 6, 2–9 (2016).
- Li, M. et al. Shape-based separation of microalga Euglena gracilis using inertial microfluidics. Sci. Rep. 7, 1–8 (2017).
- Hiramatsu, K. et al. Large-scale label-free single-cell analysis of paramylon in Euglena gracilis by high-throughput broadband Raman flow cytometry. Biomed. Opt. Express 11, 1752 (2020).
- Nitta, N. et al. Intelligent Image-Activated Cell Sorting. Cell 175, 266-276.e13 (2018).
